Figure 1. Ligands, cofactors, and non-standard residues
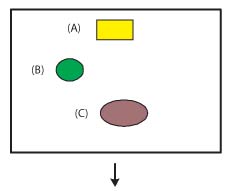
Figure 2. Molecular entity
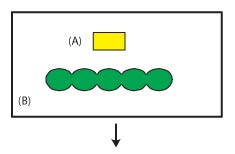
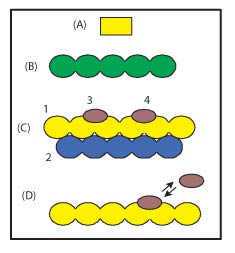
Figure 3. Molecular assembly
Introduction
A BMRB deposition requires a description of the molecular system used to collect the
reported data. The molecular system is described in a hierarchal manner starting with
any non-standard residues and/or small molecules (chemical components) in the system.
Next, the molecular entities that exist in the system and are in the sample (biopolymers
and non-polymers, including metal ions, cofactors and other small molecules) are described.
Finally, each instance of a molecular entity in the molecular system is given a name
(i.e., polypeptide one and polypeptide two for a homodimer or strand one and strand two
for a double stranded DNA system) and any unique characteristics provided. The three
levels of the molecular system hierarchy are described in more detail below.
Ligands, cofactors, and non-standard residues (chemical components)
Ligands, cofactors, and non-standard residues are any non-polymer from a simple
metal ion to a relatively complex molecule like a steriod, heme, or drug. These chemical
components may represent either chemically complete molecules (Figure 1A) or chemical moieties
like amino acid residues (Figure 1B) that are linked together to form more complex molecules.
Ligands and cofactors must also be listed as 'Molecular entities' (Figure 2A, see below),
but non-standard residues or other chemical components that are used to build more complex
molecules are not described as molecular entities. In the ADIT-NMR section where ligands,
cofactors, and non-standard residue are described, the name(s) for the molecule and a
variety of molecular characteristics are entered. The individual atoms in the component
are listed with their names and characteristics. Each chemical bond in the molecule is
defined by the atoms involved in the bond and its type. If a description of a chemical
component can be found in the PDB Ligand Depot or the BMRB ligand library, a depositor
need only supply the name of the component and its ID and the remaining required
information will be entered in the deposition processing stage by annotators.
Molecular entity (entity)
Molecular entities are chemically unique molecules that can be either a non-polymer (Figure 2A)
or a polymer (Figure 2B). For polymers, the polymer type and residue sequence are required
information. A molecular entity may represent either a single chemical component (Figure 2A) or
two or more chemical components linked together as in a protein, nucleic acid polymer,
or carbohydrate (Figure 2B).
Molecular assembly (assembly)
Molecular assemblies are composed of one or more molecular entities. A BMRB entry
contains data obtained from one molecular assembly represented by the molecules
(molecular entities) under study that are present in the sample tube. Many different
types of molecular assemblies are possible, including small non-polymeric molecules (Figure 3A),
single chain polymers (Figure 3B), homo or hetero multimeric biopolymer complexes possibly with
ligands (Figure 3C), complexes in very slow to fast exchange with free ligands (Figure 3D). If
the molecular assembly represents more than one molecule, the molecules may form either
a tight or loose complex. A complete list of the molecules that make up the assembly is
required. For molecular assembly shown in Figure 3C, the assembly molecule list
contains four items even though two of the molecules (magenta) are chemically identical.
The presence of any intra- or inter-molecular crosslinking bonds also needs to be reported.
Example molecular assemblies with instructions for entering a deposition
1) Ribonuclease (single chain polypeptide without ligands, Figure 3B):
Step 1) The Ligand section can be ignored
Step 2) One Molecular entity section must be completed. In this section at the prompt 'Brief molecule name ...' enter 'Rnase'
and then complete the remaining fields as required.
Step 3) The Molecular assembly section must be completed, but only one column in the molecular assembly
contents table needs to be filled in. In the molecular assembly contents table, for the
prompt 'Assembly component name' enter 'Rnase' (the value should be in the pull-down list)
and then complete the remaining fields in the column and the section as required.
2) Human zinc finger 2HX1 (single chain polypeptide with two zinc ligands):
Step 1) In the Entry interview for the prompt 'The system contains a metal and/or a non-polymeric organic ligand' check 'yes'.
Step 2) One ligand section must be completed. At the prompt 'Ligand or chemical component name' enter 'zinc'. Continue to
answer the remaining mandatory prompts.
Step 3) Two Molecular entity sections must be completed, one for zinc and one for the protein. In the Molecular entity section,
click on the 'Replicate' button to create a second Molecular entity form. In the second form, for the prompt 'Brief molecule name ...' enter 'zinc' ('zinc' should appear in the
pull-down list). For 'Molecule type' enter 'non-polymer' from the pull-down list. Next, click on the 'Save Changes' button as this will reset the form
with the fields appropriate for a non-polymer molecular entity. Complete the remaining mandatory fields in the form and enter any additional information
that you would like. Click the 'Save Changes' button.
Step 4) Select the first Molecular entity section in the list and at the prompt 'Brief molecular name ...' enter something like '2HX1 polypeptide'.
Complete the remaining mandatory fields in the form and any additional fields that are relevant.
Step 5) Select the Molecular assembly section. For the prompt 'Molecular assembly name', enter something like '2HX1'. Enter '3' for the total number of
biopolymers and ligands. At the prompt 'Are there cross-linking or other intermolecular bonds ...', click 'yes' and then click on the 'Save Changes' button.
Step 6) In the 'Molecular assembly contents' table, three columns need to be filled out. For the prompt 'Assembly component name', the values '2HX1 polypeptide',
zinc 1, and zinc 2 would be appropriate. For the prompt 'Molecular entity name to assign ...', the appropriate values can be selected from the pull-down list,
'2HX1 polypeptide' for the component '2HX1 polypeptide' and 'zinc' for the components 'zinc 1' and 'zinc 2'. Complete the remaining required fields in the table.
Step 7) For each bond between the polypeptide and a ligated zinc a column in the 'Intermolecular and interresidue bonds ...' table must be completed.
Step 8) If atoms are deleted in forming intermolecular bonds, the deleted atoms need to be reported in the 'Atoms deleted ...' table.
3) Hemoglobin (multichain, multiligand complex):
Step 1) In the Entry interview for the prompt 'The system contains a metal and/or a non-polymeric organic ligand' check 'yes'.
Step 2) One ligand section must be completed. At the prompt 'Ligand or chemical component name' enter 'heme'. Continue to
answer the remaining mandatory prompts.
Step 3) Three Molecular entity sections must be completed, one for heme and one for each type of polypeptide chain (A and B) in the complex. In the Molecular entity section,
click on the 'Replicate' button twice to create a total of three Molecular entity forms. In the last form, for the prompt 'Brief molecule name ...' enter 'heme' ('heme' should appear in the
pull-down list). For 'Molecule type' enter 'non-polymer' from the pull-down list. Next, click on the 'Save Changes' button as this will reset the form
with the fields appropriate for a non-polymer molecular entity. Complete the remaining mandatory fields in the form and enter any additional information
that you would like. Click the 'Save Changes' button.
Step 4) Select the first Molecular entity section in the list and at the prompt 'Brief molecular name ...' enter something like 'Alpha chain'.
Complete the remaining mandatory fields in the form and any additional fields that are relevant.
Step 5) Select the second Molecular entity section in the list and at the prompt 'Brief molecular name ...' enter something like 'Beta chain'.
Complete the remaining mandatory fields in the form and any additional fields that are relevant.
Step 6) Select the Molecular assembly section. For the prompt 'Molecular assembly name', enter something like 'hemoglobin'. Enter '8' for the total number of
biopolymers and ligands. At the prompt 'Are there cross-linking or other intermolecular bonds ...', click 'yes' and then click on the 'Save Changes' button.
Step 7) In the 'Molecular assembly contents' table, eight columns need to be filled out. For the prompt 'Assembly component name', the values 'alpha chain 1', 'alpha chain 2', 'beta chain 1',
'beta chain 2', 'heme 1', 'heme 2', 'heme 3', 'heme 4' would be appropriate. For the prompt 'Molecular entity name to assign ...', the appropriate values can be selected from the pull-down list,
'Alpha chain' for the components 'alpha chain 1' and 'alpha chain 2', 'Beta chain' for the components 'beta chain 1' and beta chain 2' and 'Heme' for the components 'heme 1', 'heme 2',
'heme 3', and 'heme 4'. Complete the remaining required fields in the table.
Step 8) For each bond between the polypeptide and a ligated heme a column in the 'Intermolecular and interresidue bonds ...' table must be completed.
Step 9) If atoms are deleted in forming intermolecular bonds, the deleted atoms need to be reported in the 'Atoms deleted ...' table.
 Figure 2. Molecular entity
Figure 2. Molecular entity
 Figure 3. Molecular assembly
Figure 3. Molecular assembly

 Figure 2. Molecular entity
Figure 2. Molecular entity
 Figure 3. Molecular assembly
Figure 3. Molecular assembly
