BMRB Entry 19654
Click here to enlarge.
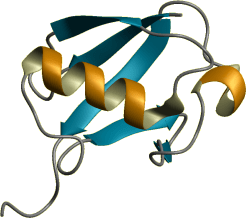
PDB ID: 2mhs
Entry in NMR Restraints Grid
Validation report in NRG-CING
Chem Shift validation: AVS_full, LACS, SPARTA
BMRB Entry DOI: doi:10.13018/BMR19654
MolProbity Validation Chart
NMR-STAR file interactive viewer.
NMR-STAR v3 text file.
NMR-STAR v2.1 text file (deprecated)
XML gzip file.
RDF gzip file.
All files associated with the entry
Title: NMR Structure of human Mcl-1 PubMed: 24789074
Deposition date: 2013-12-04 Original release date: 2014-05-12
Authors: Liu, Gaohua; Poppe, Leszek; Aoki, Ken; Yamane, Harvey; Lewis, Jeffrey; Szyperski, Thomas
Citation: Liu, Gaohua; Poppe, Leszek; Aoki, Ken; Yamane, Harvey; Lewis, Jeffrey; Szyperski, Thomas. "High-quality NMR Structure of Human Anti-apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design" PLOS 9, e96521-e96521 (2014).
Assembly members:
entity, polymer, 328 residues, 37715.047 Da.
Natural source: Common Name: Human Taxonomy ID: 9606 Superkingdom: Eukaryota Kingdom: Metazoa Genus/species: Homo sapiens
Experimental source: Production method: recombinant technology Host organism: Escherichia coli
- assigned_chemical_shifts
| Data type | Count |
| 13C chemical shifts | 548 |
| 15N chemical shifts | 175 |
| 1H chemical shifts | 1153 |
Additional metadata:
Download simulated HSQC data in one of the following formats:
CSV: Backbone
or all simulated shifts
SPARKY: Backbone
or all simulated shifts