BMRB Entry 30204
Click here to enlarge.
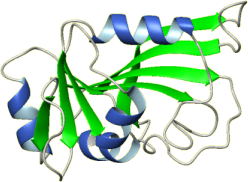
PDB ID: 5tx8
Entry in NMR Restraints Grid
Validation report in NRG-CING
Chem Shift validation: AVS_full
BMRB Entry DOI: doi:10.13018/BMR30204
MolProbity Validation Chart
NMR-STAR file interactive viewer.
NMR-STAR v3 text file.
NMR-STAR v2.1 text file (deprecated)
XML gzip file.
RDF gzip file.
All files associated with the entry
Title: Solution structure of the de novo mini protein gHH_44
Deposition date: 2016-11-15 Original release date: 2017-09-25
Authors: Buchko, G.; Bahl, C.; Baker, D.
Citation: Bahl, C.; Buchko, G.; Baker, D.. "Biophysical characterization of hyperstable constrained peptides." . ., .-..
Assembly members:
entity_1, polymer, 28 residues, 3366.862 Da.
Natural source: Common Name: not available Taxonomy ID: 32630 Superkingdom: not available Kingdom: not available Genus/species: not available not available
Experimental source: Production method: recombinant technology Host organism: Escherichia coli
Entity Sequences (FASTA):
entity_1: AEDCERIRKELEKNPNDEIK
KKLEKCQA
- assigned_chemical_shifts
- spectral_peak_list
| Data type | Count |
| 13C chemical shifts | 117 |
| 15N chemical shifts | 29 |
| 1H chemical shifts | 164 |
Additional metadata:
Download simulated HSQC data in one of the following formats:
CSV: Backbone
or all simulated shifts
SPARKY: Backbone
or all simulated shifts